Содержание
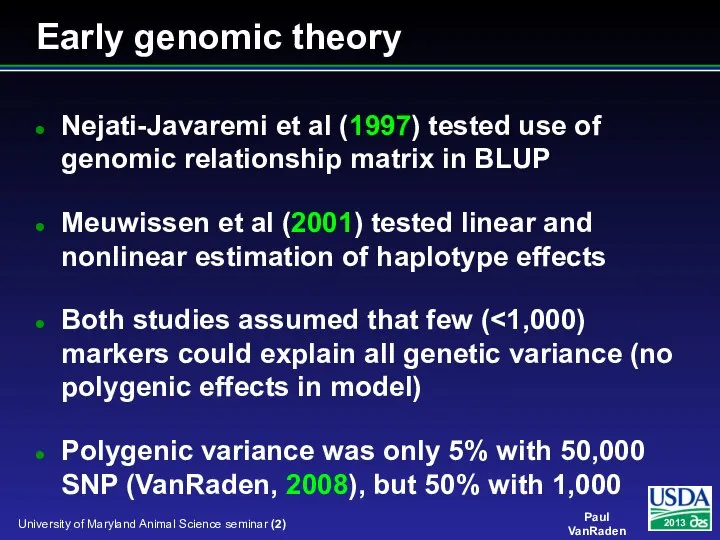
- 2. Early genomic theory Nejati-Javaremi et al (1997) tested use of genomic relationship matrix in BLUP Meuwissen
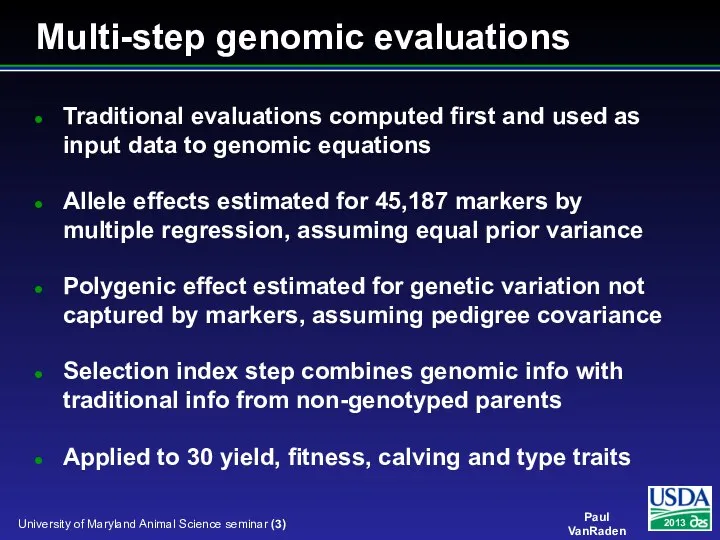
- 3. Multi-step genomic evaluations Traditional evaluations computed first and used as input data to genomic equations Allele
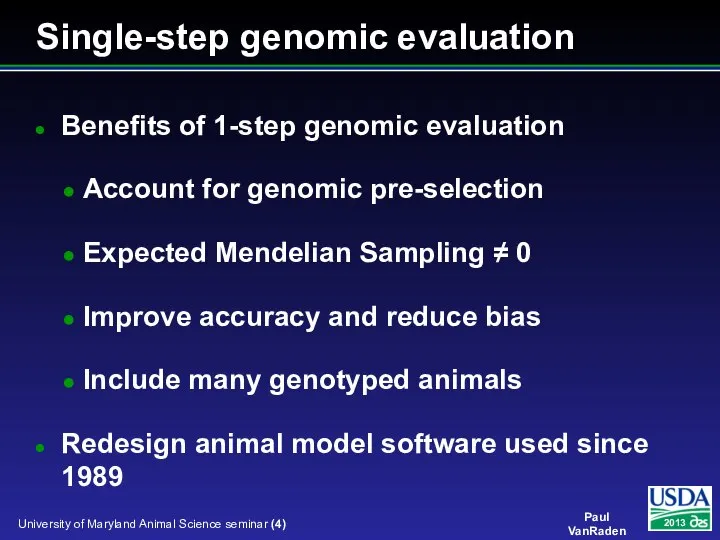
- 4. Benefits of 1-step genomic evaluation Account for genomic pre-selection Expected Mendelian Sampling ≠ 0 Improve accuracy
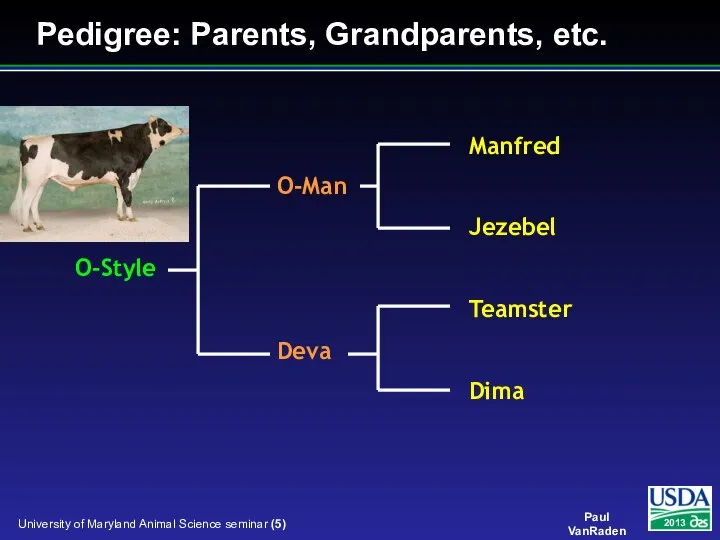
- 5. Pedigree: Parents, Grandparents, etc.
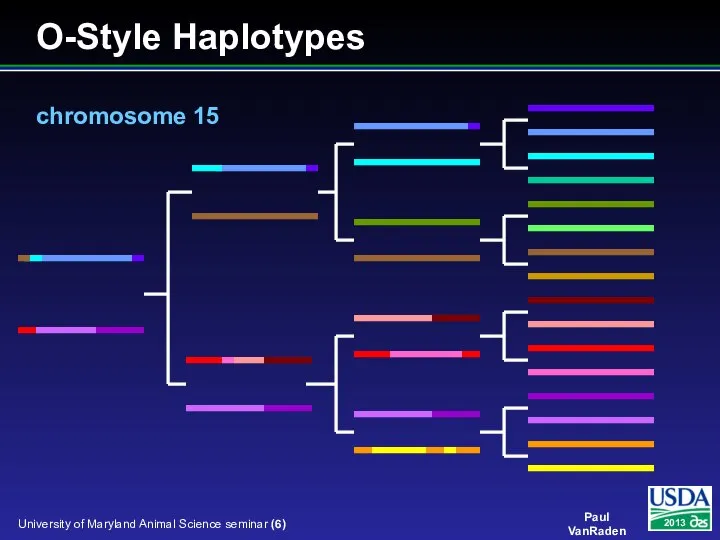
- 6. O-Style Haplotypes chromosome 15
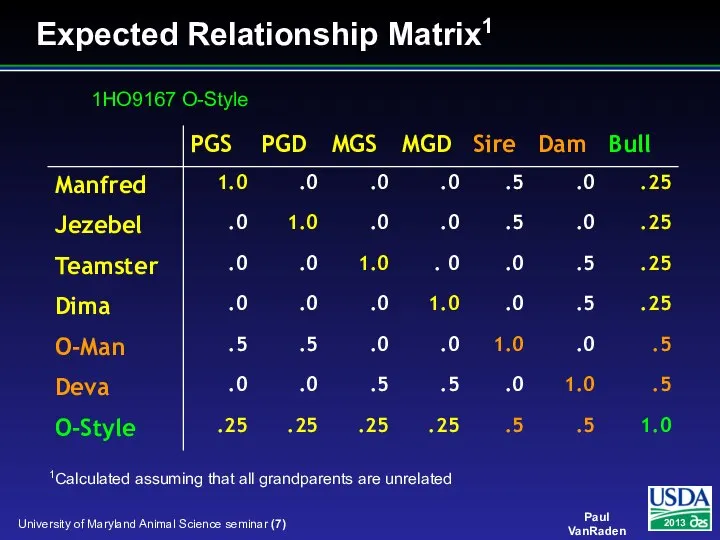
- 7. Expected Relationship Matrix1 1Calculated assuming that all grandparents are unrelated 1HO9167 O-Style
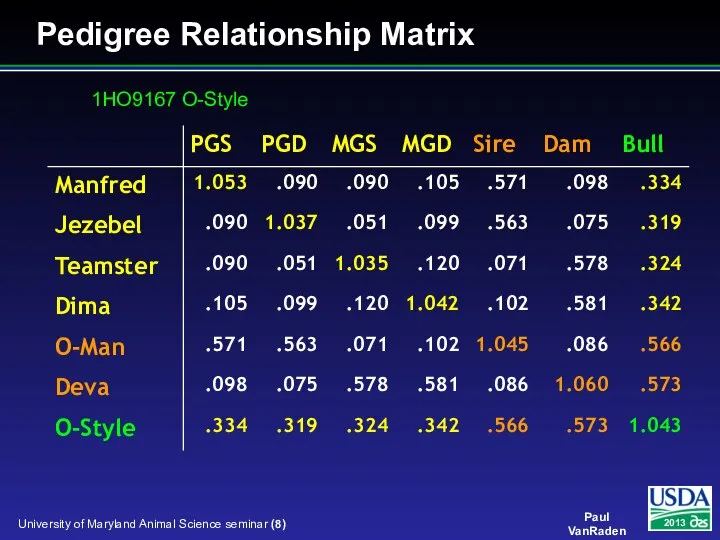
- 8. Pedigree Relationship Matrix 1HO9167 O-Style
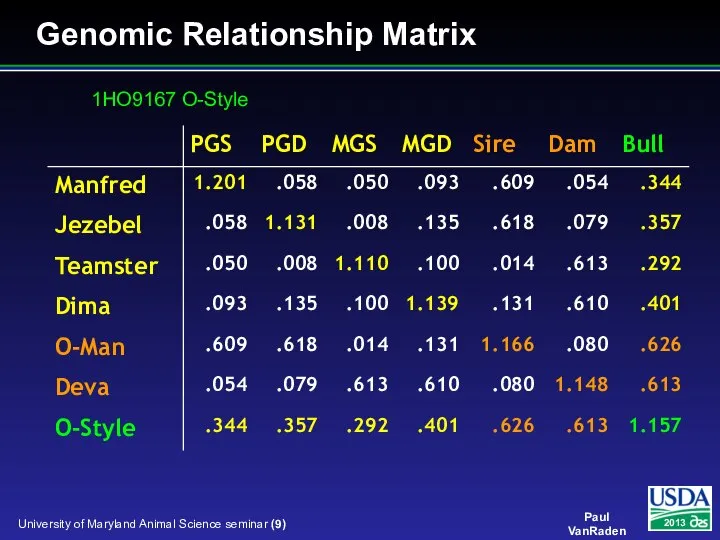
- 9. Genomic Relationship Matrix 1HO9167 O-Style
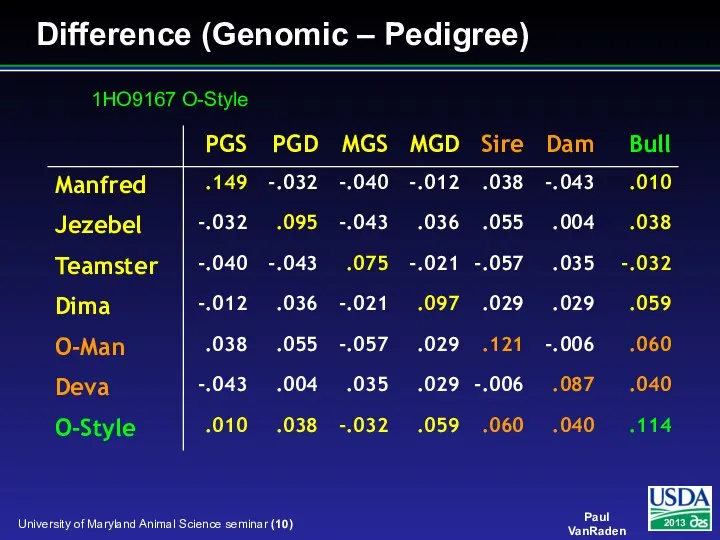
- 10. Difference (Genomic – Pedigree) 1HO9167 O-Style
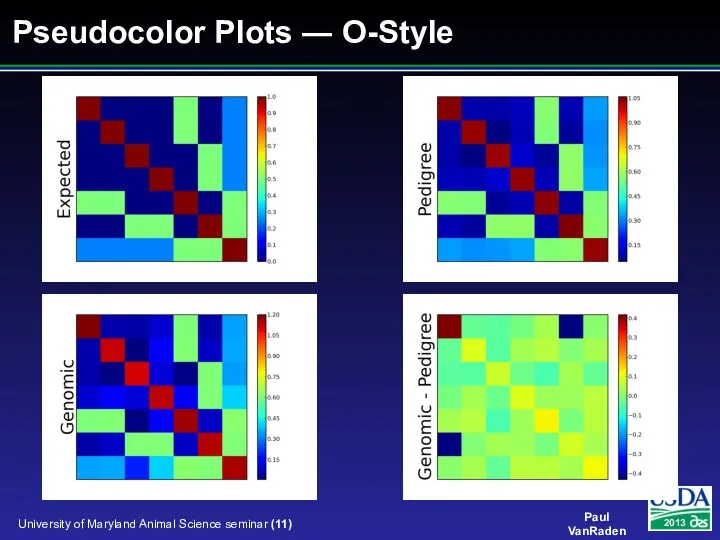
- 11. Pseudocolor Plots ― O-Style
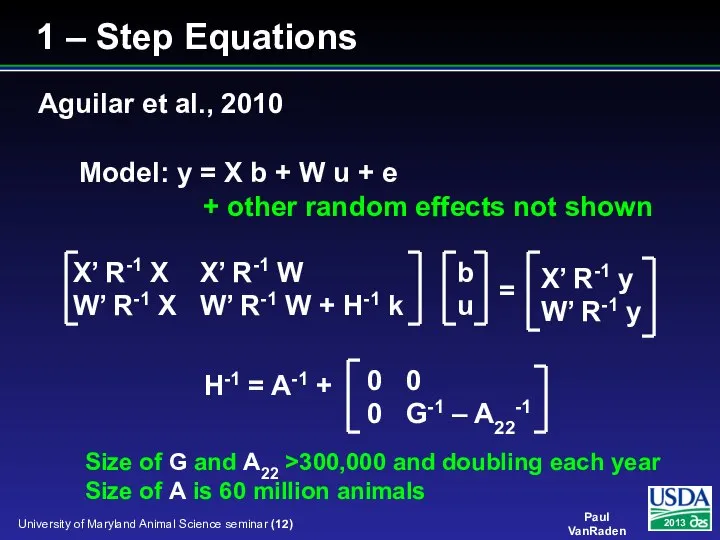
- 12. X’ R-1 X X’ R-1 W W’ R-1 X W’ R-1 W + H-1 k Model:
- 13. X’R-1X X’R-1W 0 0 W’R-1X W’R-1W+A-1k Q Q 0 Q’ -G/k 0 0 Q’ 0 A22/k
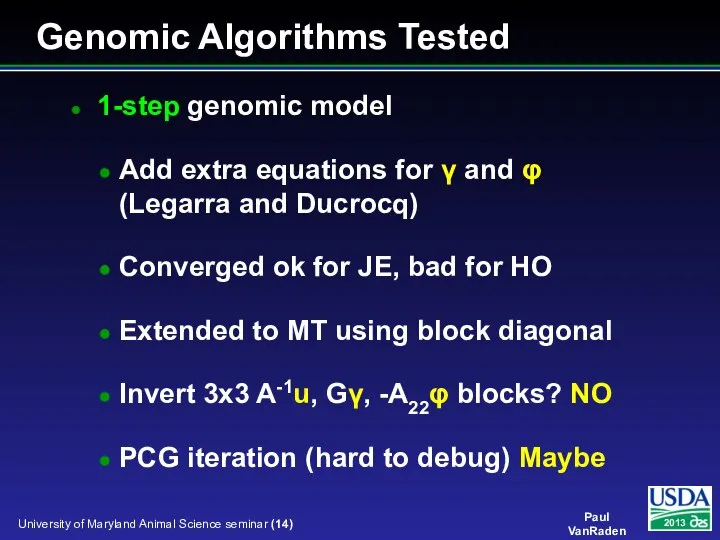
- 14. 1-step genomic model Add extra equations for γ and φ (Legarra and Ducrocq) Converged ok for
- 15. Multi-step insertion of GEBV [W’R-1W + A-1k] u = W’R-1y (without G) Previous studies added genomic
- 16. National U.S. Jersey data 4.4 million lactation phenotypes 4.1 million animals in pedigree Multi-trait milk, fat,
- 17. Jersey Results New = 1-step GPTA milk, Old = multi-step GPTA milk
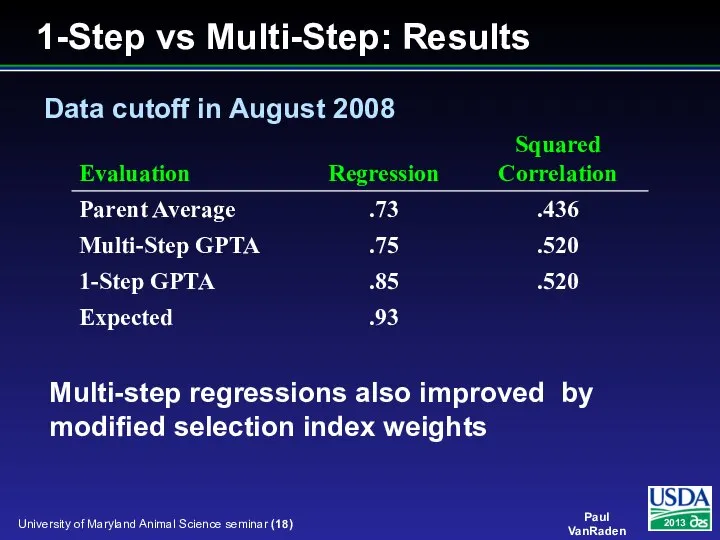
- 18. Multi-step regressions also improved by modified selection index weights Data cutoff in August 2008 1-Step vs
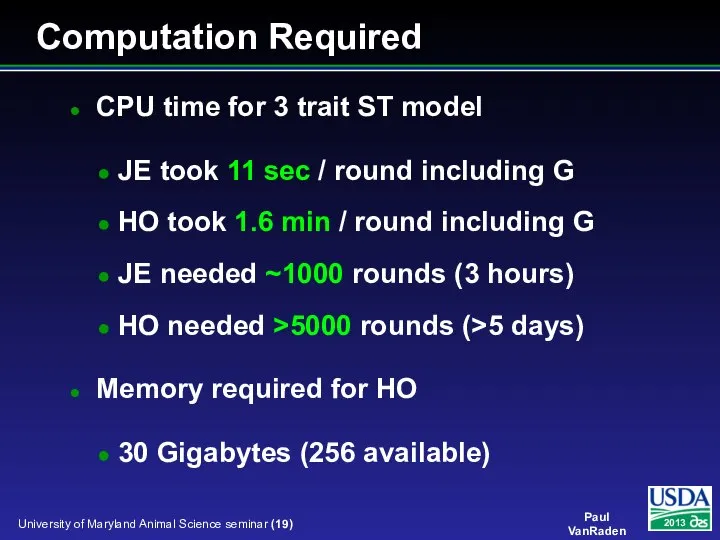
- 19. CPU time for 3 trait ST model JE took 11 sec / round including G HO
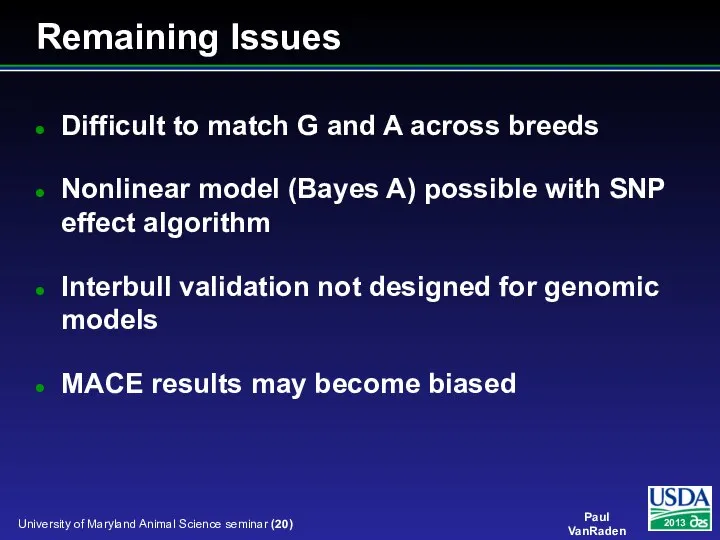
- 20. Difficult to match G and A across breeds Nonlinear model (Bayes A) possible with SNP effect
- 21. Steps to prepare genotypes Nominate animal for genotyping Collect blood, hair, semen, nasal swab, or ear
- 22. Ancestor Validation and Discovery Ancestor discovery can accurately confirm, correct, or discover parents and more distant
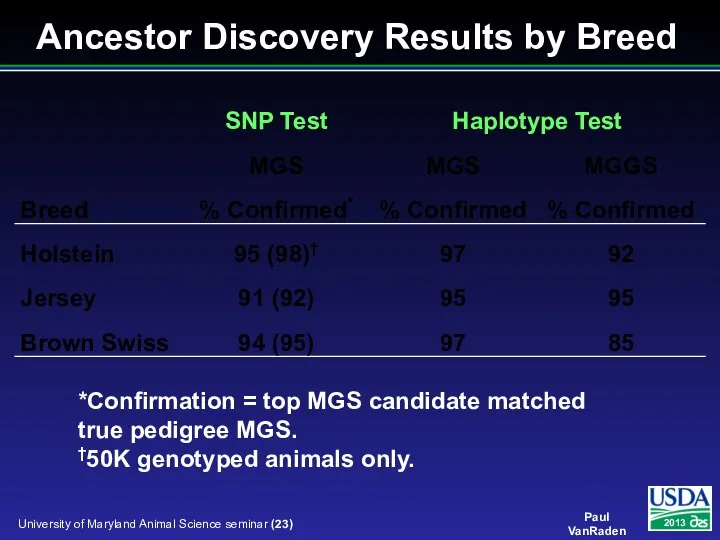
- 23. Ancestor Discovery Results by Breed *Confirmation = top MGS candidate matched true pedigree MGS. †50K genotyped
- 24. One step model includes: 72 million lactation phenotypes 50 million animals in pedigree 29 million permanent
- 25. Model options now include: Multi-trait models Multiple class and regress variables Suppress some factors / each
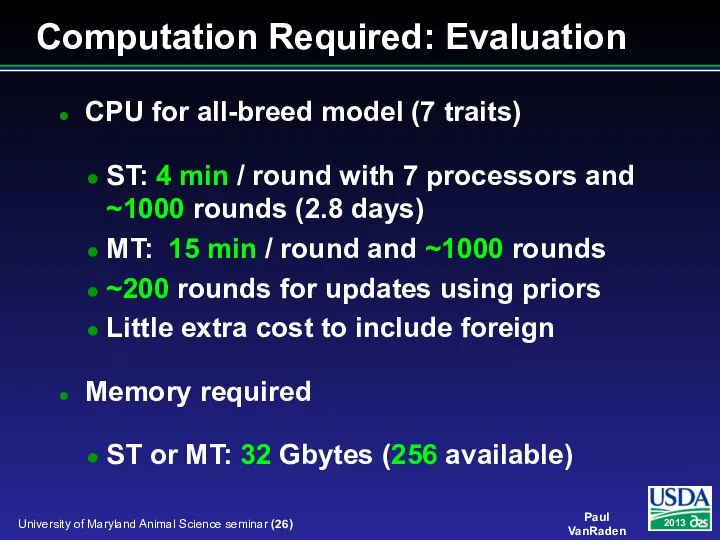
- 26. CPU for all-breed model (7 traits) ST: 4 min / round with 7 processors and ~1000
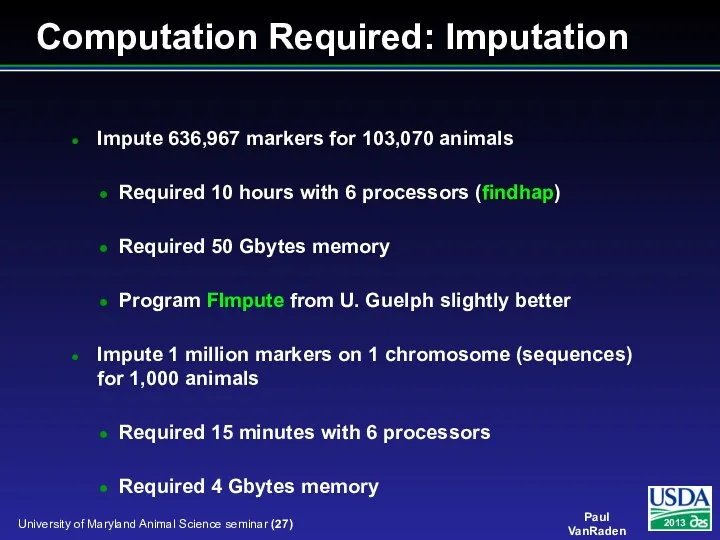
- 27. Impute 636,967 markers for 103,070 animals Required 10 hours with 6 processors (findhap) Required 50 Gbytes
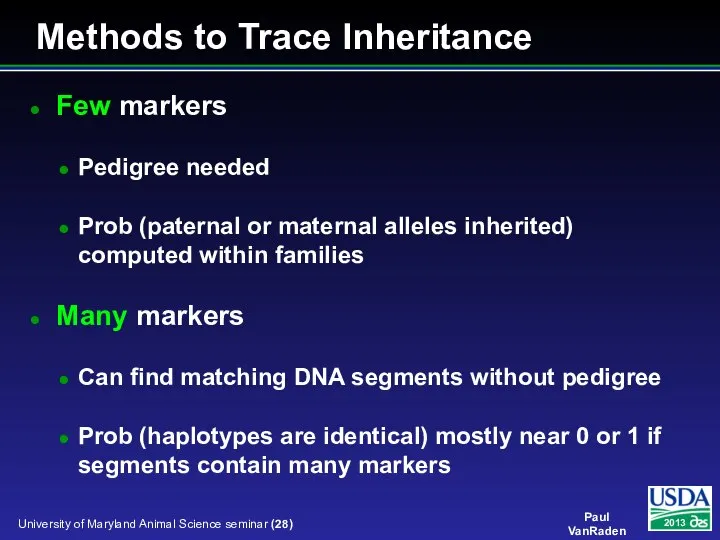
- 28. Methods to Trace Inheritance Few markers Pedigree needed Prob (paternal or maternal alleles inherited) computed within
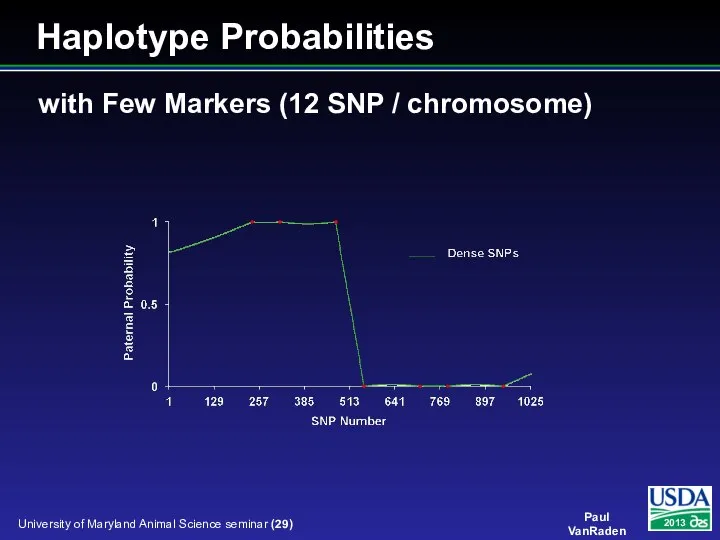
- 29. with Few Markers (12 SNP / chromosome) Haplotype Probabilities
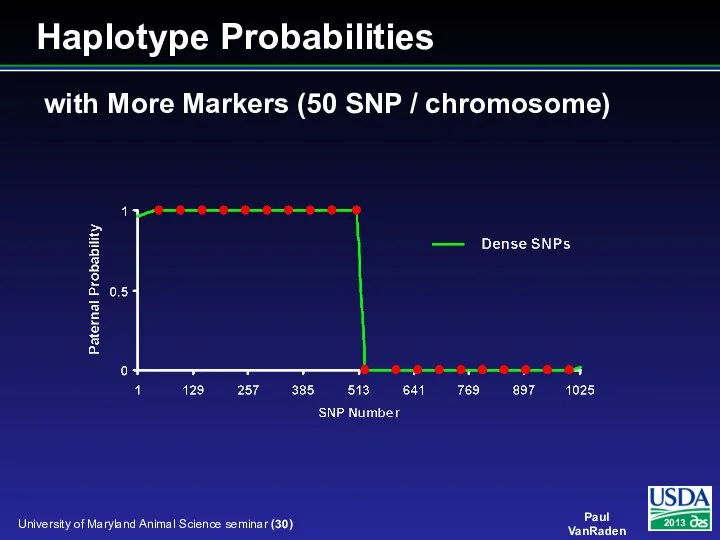
- 30. with More Markers (50 SNP / chromosome) Haplotype Probabilities
- 31. Haplotyping Program: findhap.f90 Population haplotyping Divide chromosomes into segments List haplotypes by genotype match Similar to
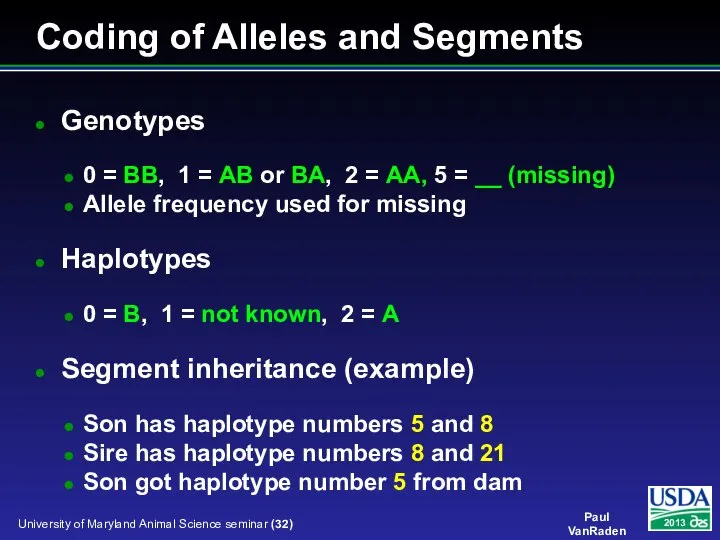
- 32. Coding of Alleles and Segments Genotypes 0 = BB, 1 = AB or BA, 2 =
- 33. Population Haplotyping Steps Put first genotype into haplotype list Check next genotype against list Do any
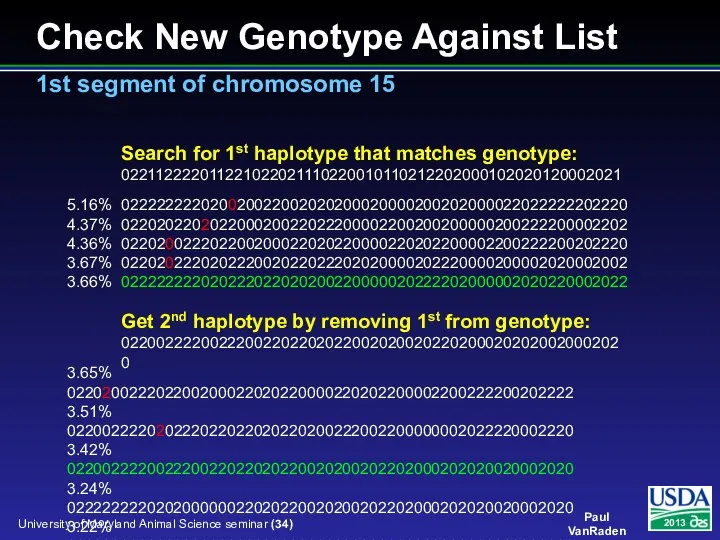
- 34. Check New Genotype Against List 1st segment of chromosome 15 5.16% 022222222020020022002020200020000200202000022022222202220 4.37% 022020220202200020022022200002200200200000200222200002202 4.36% 022020022202200200022020220000220202200002200222200202220
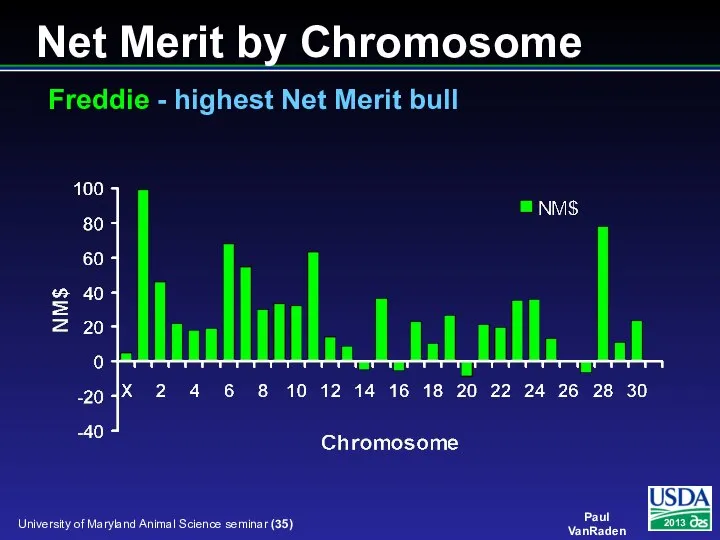
- 35. Net Merit by Chromosome Freddie - highest Net Merit bull
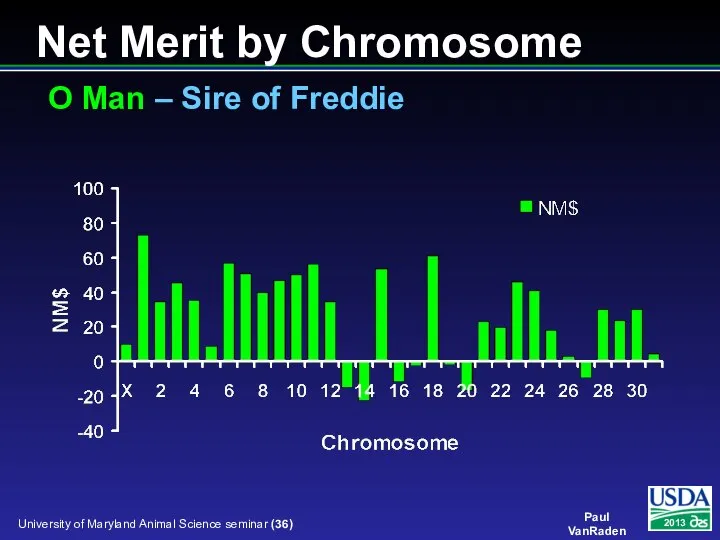
- 36. Net Merit by Chromosome O Man – Sire of Freddie
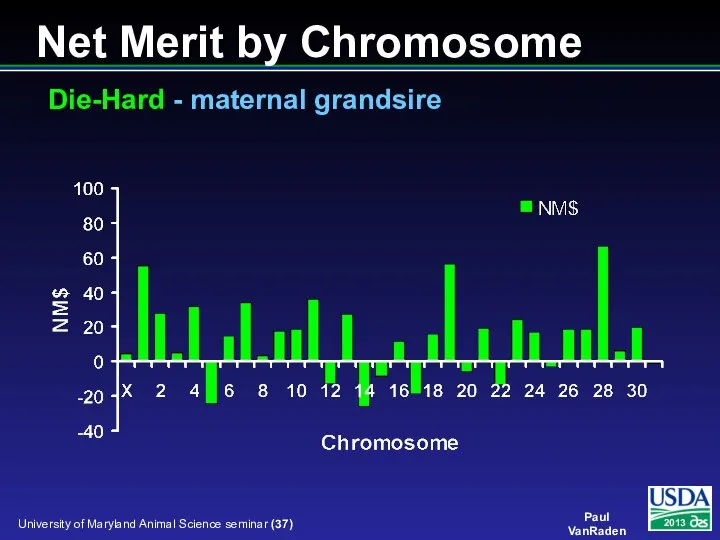
- 37. Net Merit by Chromosome Die-Hard - maternal grandsire
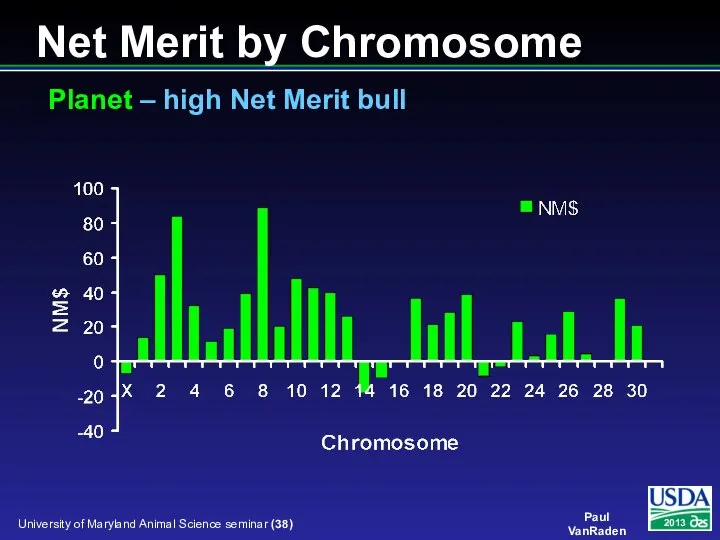
- 38. Net Merit by Chromosome Planet – high Net Merit bull
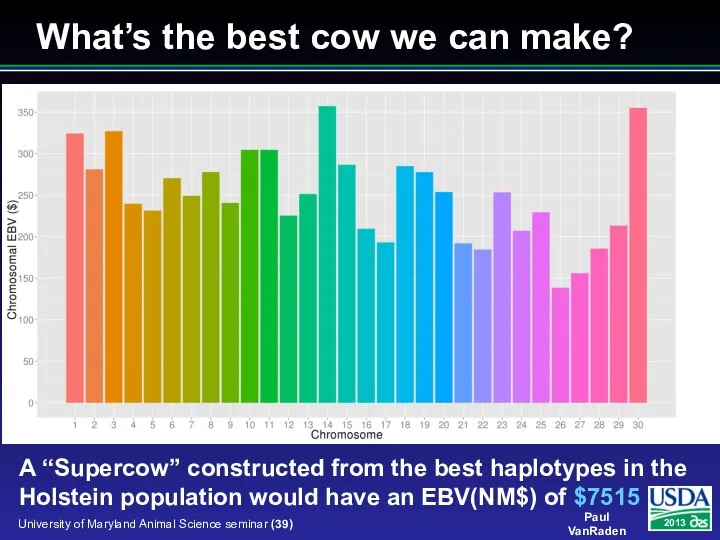
- 39. What’s the best cow we can make? A “Supercow” constructed from the best haplotypes in the
- 40. Conclusions 1-step genomic evaluations tested Inversion avoided using extra equations Converged well for JE but not
- 41. Conclusions Foreign data can add to national evaluations In one step model instead of post-process High
- 43. Скачать презентацию

![Multi-step insertion of GEBV [W’R-1W + A-1k] u = W’R-1y (without G)](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/1140365/slide-14.jpg)










 Фотосинтез
Фотосинтез Сукцессия. Саморазвитие экосистемы
Сукцессия. Саморазвитие экосистемы Невидимые нити в осеннем лесу
Невидимые нити в осеннем лесу Брахиозавр (Плечистый Ящер)
Брахиозавр (Плечистый Ящер) Царства живой природы
Царства живой природы Способы размножения растений. Урок №24
Способы размножения растений. Урок №24 Презентация на тему ТИП ПЛОСКИЕ ЧЕРВИ
Презентация на тему ТИП ПЛОСКИЕ ЧЕРВИ  Викторина по биологии
Викторина по биологии Адаптация. Ее формы и механизмы
Адаптация. Ее формы и механизмы Тест по теме Свиньи
Тест по теме Свиньи Видообразование
Видообразование Биологияны оқыту әдістері мен әдістемелік тәсілдері
Биологияны оқыту әдістері мен әдістемелік тәсілдері Презентация на тему Медузы
Презентация на тему Медузы  Миоглобин. Тропонин. Церулоплазмин. Гаптоглобин
Миоглобин. Тропонин. Церулоплазмин. Гаптоглобин Уровни организации. Структура тела. Органы и системы органов
Уровни организации. Структура тела. Органы и системы органов Vylučovanie - močová sústava človeka
Vylučovanie - močová sústava človeka Вітамін А
Вітамін А Строение белков
Строение белков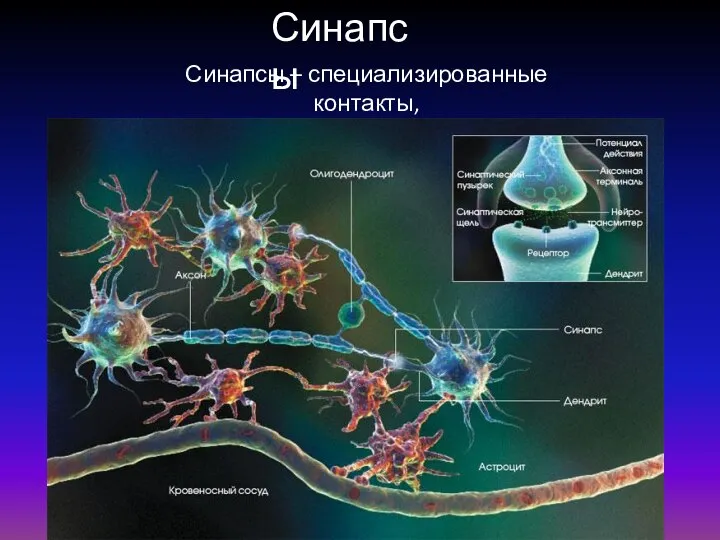 Синапсы. Классификация синапсов
Синапсы. Классификация синапсов Презентация на тему НАДКЛАСС РЫБЫ
Презентация на тему НАДКЛАСС РЫБЫ  Половая система
Половая система Высшие споровые растения, происхождение, общая характеристика. Жизненный цикл высших споровых растений
Высшие споровые растения, происхождение, общая характеристика. Жизненный цикл высших споровых растений Печеночный сосальщик. Цикл развития
Печеночный сосальщик. Цикл развития Анастомоз. Анатомия
Анастомоз. Анатомия Сравнительная шкала разрешающей способности невооруженного глаза, светового и электронного микрскопов
Сравнительная шкала разрешающей способности невооруженного глаза, светового и электронного микрскопов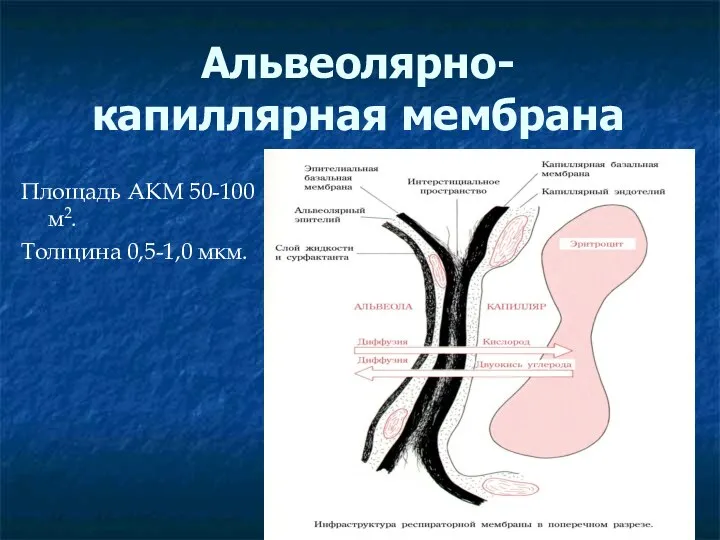 Альвеолярно-капиллярная мембрана
Альвеолярно-капиллярная мембрана Тип Моллюски
Тип Моллюски Развитие насекомых
Развитие насекомых