Содержание
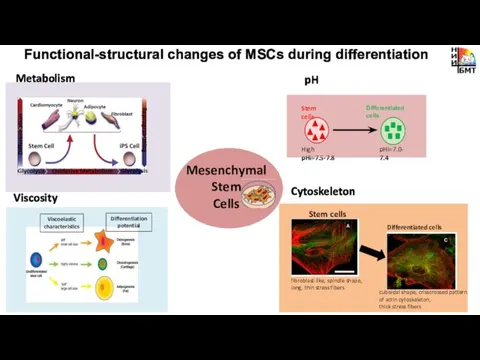
- 2. Metabolism pH Cytoskeleton Functional-structural changes of MSCs during differentiation Viscosity Mesenchymal Stem Cells
- 3. Effective control of MSCs differentiation - great challenge Complex analysis is required!!!
- 4. Methods of the stem cells morphology and physiology investigation Feature Method Cell markers • Flow cytometry
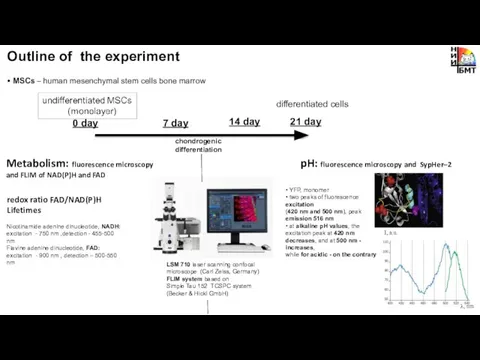
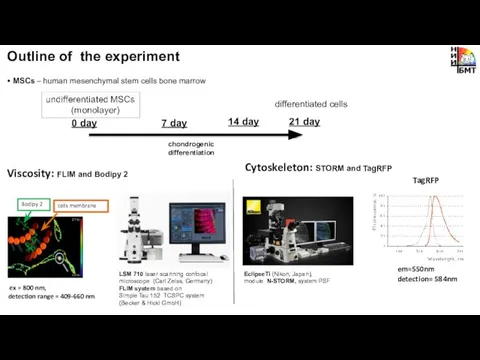
- 5. Outline of the experiment • MSCs – human mesenchymal stem cells bone marrow Metabolism: fluorescence microscopy
- 6. • MSCs – human mesenchymal stem cells bone marrow LSM 710 laser scanning confocal microscope (Carl
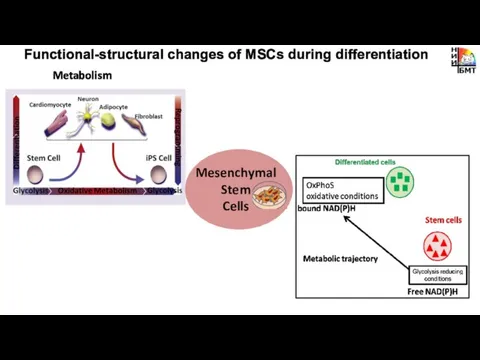
- 7. Metabolism Functional-structural changes of MSCs during differentiation Mesenchymal Stem Cells
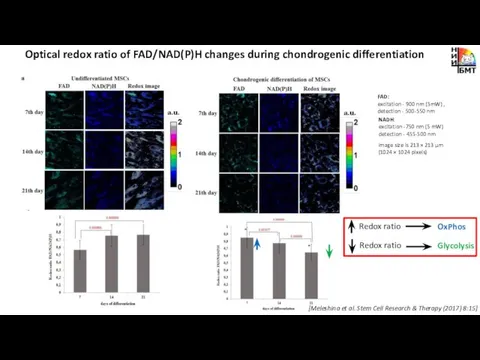
- 8. Optical redox ratio of FAD/NAD(P)H changes during chondrogenic differentiation NADH: excitation -750 nm (5 mW) detection
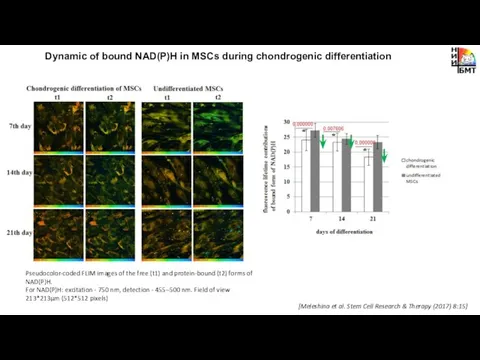
- 9. Dynamic of bound NAD(P)H in MSCs during chondrogenic differentiation [Meleshina et al. Stem Cell Research &
- 10. pH Functional-structural changes of MSCs during differentiation Mesenchymal Stem Cells
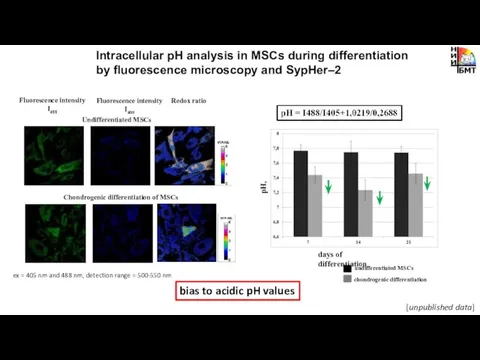
- 11. Intracellular pH analysis in MSCs during differentiation by fluorescence microscopy and SypHer–2 days of differentiation pH,
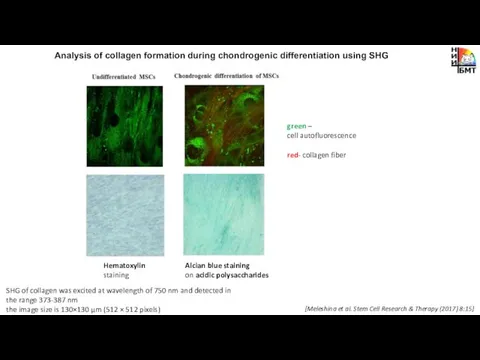
- 12. Analysis of collagen formation during chondrogenic differentiation using SHG green – cell autofluorescence red- collagen fiber
- 13. Cytoskeleton Functional-structural changes of MSCs during differentiation Viscosity Mesenchymal Stem Cells
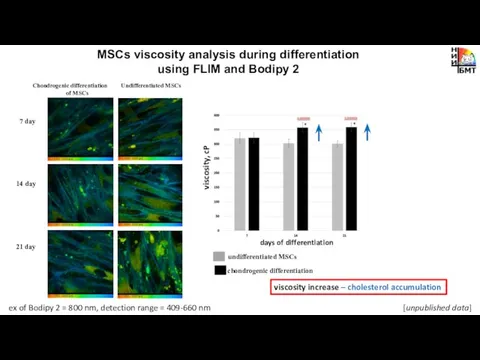
- 14. MSCs viscosity analysis during differentiation using FLIM and Bodipy 2 chondrogenic differentiation undifferentiated MSCs viscosity increase
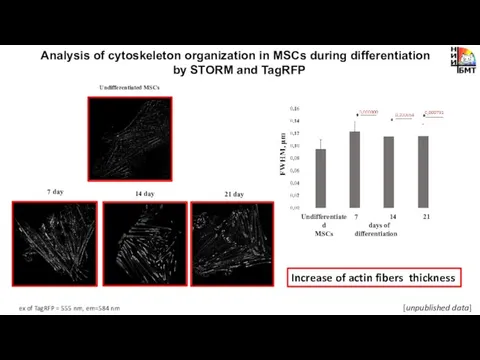
- 15. Analysis of cytoskeleton organization in MSCs during differentiation by STORM and TagRFP Undifferentiated MSCs 7 day
- 16. take home message Metabolic plasticity of MSCs during chondrogenic differentiation: glycolysis – more glycolytic state Intracellular
- 17. Acknowledgements This work has been financially supported by Russian Science Foundation (grants No. 14-15-00536) M.V. Shirmanova
- 19. Скачать презентацию




 Ребятам о муравьях (средняя группа)
Ребятам о муравьях (средняя группа) Растения луга
Растения луга Что растёт на клумбе
Что растёт на клумбе Остеология. Осевой скелет
Остеология. Осевой скелет Мутація у живих організмів
Мутація у живих організмів Обобщающий урок Ткани животных
Обобщающий урок Ткани животных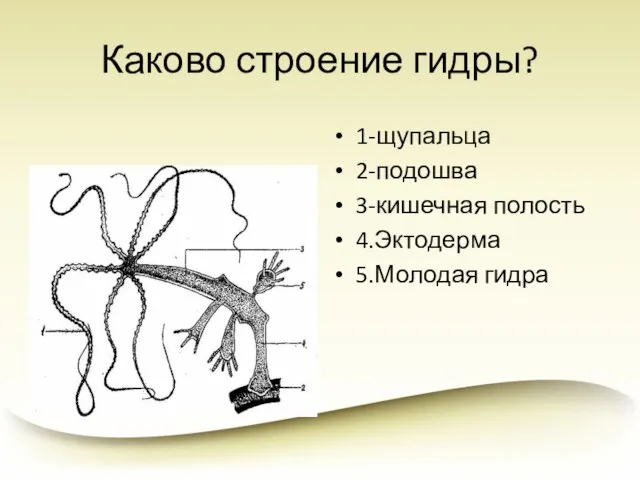 Каково строение гидры?
Каково строение гидры? Комары
Комары Єноти - хижі ссавці, що належать до однойменного сімейства єнотових
Єноти - хижі ссавці, що належать до однойменного сімейства єнотових Микробы
Микробы Деление клетки. Митоз
Деление клетки. Митоз Орг-вещ9конц
Орг-вещ9конц Запуск аквариума. Инструкция для начинающих
Запуск аквариума. Инструкция для начинающих Витамины. Классификация
Витамины. Классификация Презентация на тему Увеличительные приборы и правила работы с ними
Презентация на тему Увеличительные приборы и правила работы с ними  Пластиды
Пластиды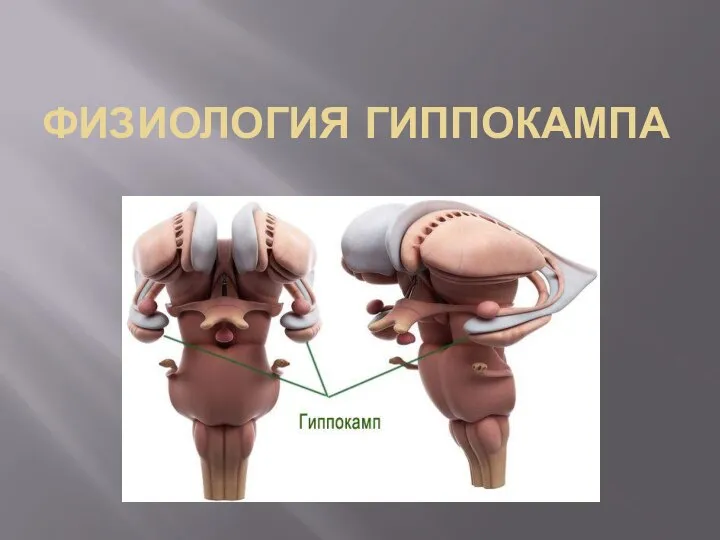 Физиология гиппокампа
Физиология гиппокампа Гиганты и лилипуты в мире насекомых
Гиганты и лилипуты в мире насекомых Критерии вида
Критерии вида Роль витаминов в жизни человека
Роль витаминов в жизни человека Пустельга обыкновенная
Пустельга обыкновенная Звуки, которые может издавать человек
Звуки, которые может издавать человек Проращивание фасоли
Проращивание фасоли Генетика 18
Генетика 18 Методи селекції
Методи селекції Мои питомцы
Мои питомцы Наглядное пособие Скелет Бронтозавра
Наглядное пособие Скелет Бронтозавра Окружающий мир (1 класс)
Окружающий мир (1 класс)