Содержание
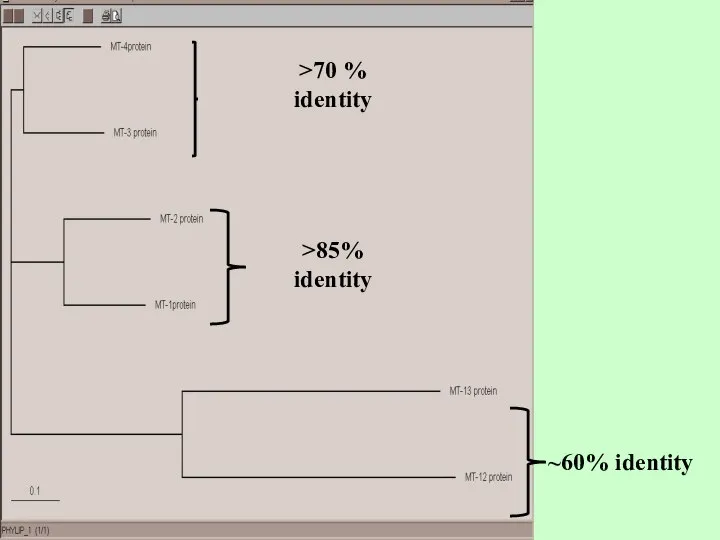
- 2. >70 % identity >85% identity ~60% identity
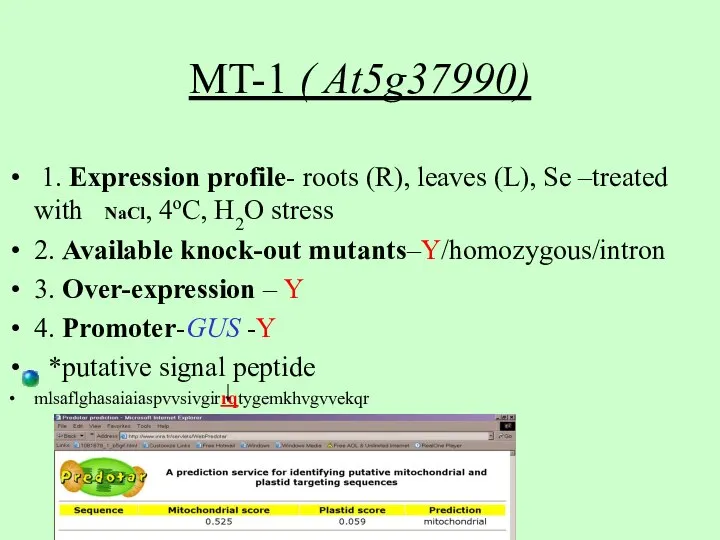
- 3. 1. Expression profile- roots (R), leaves (L), Se –treated with NaCl, 4oC, H2O stress 2. Available
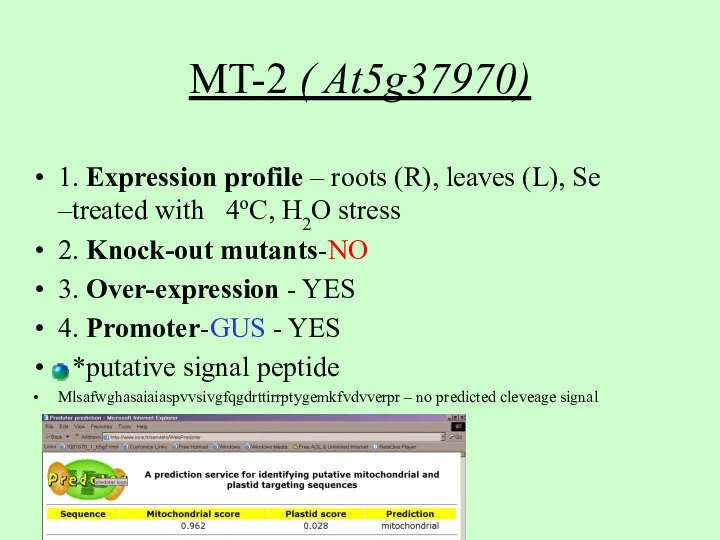
- 4. MT-2 ( At5g37970) 1. Expression profile – roots (R), leaves (L), Se –treated with 4oC, H2O
- 5. MT-3 ( At5g38780) 1. Expression profile- R, Leaves, Se, SeNaCl, Se4oC, Water treated leaves 2. Available
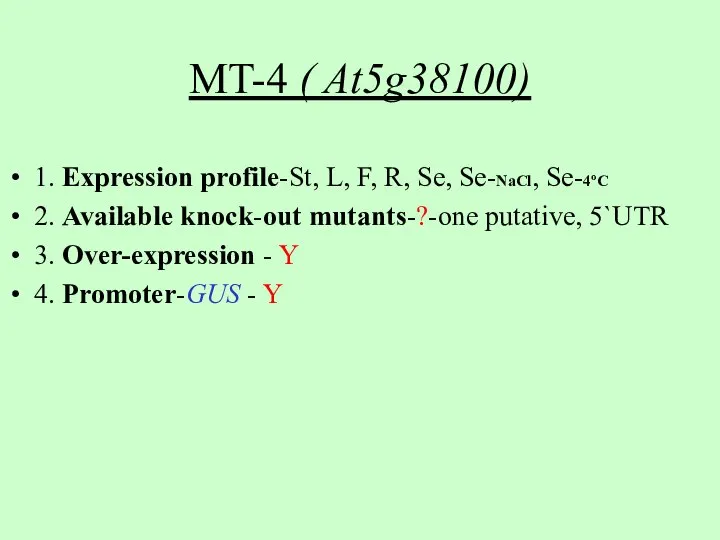
- 6. MT-4 ( At5g38100) 1. Expression profile-St, L, F, R, Se, Se-NaCl, Se-4oC 2. Available knock-out mutants-?-one
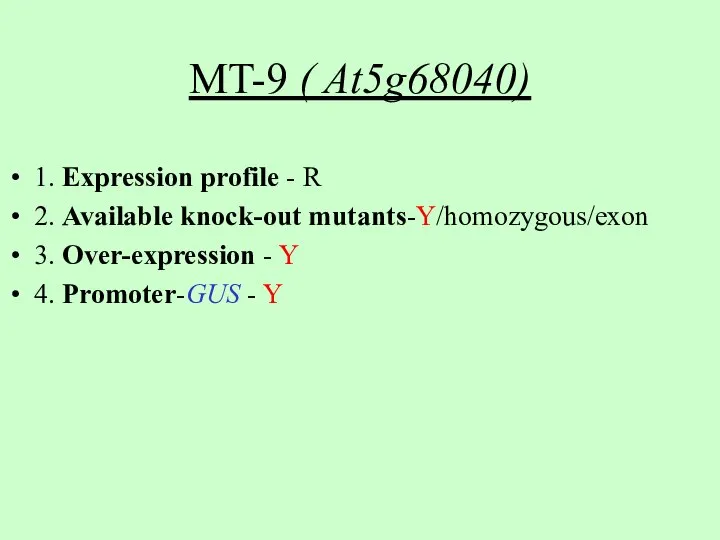
- 7. MT-9 ( At5g68040) 1. Expression profile - R 2. Available knock-out mutants-Y/homozygous/exon 3. Over-expression - Y
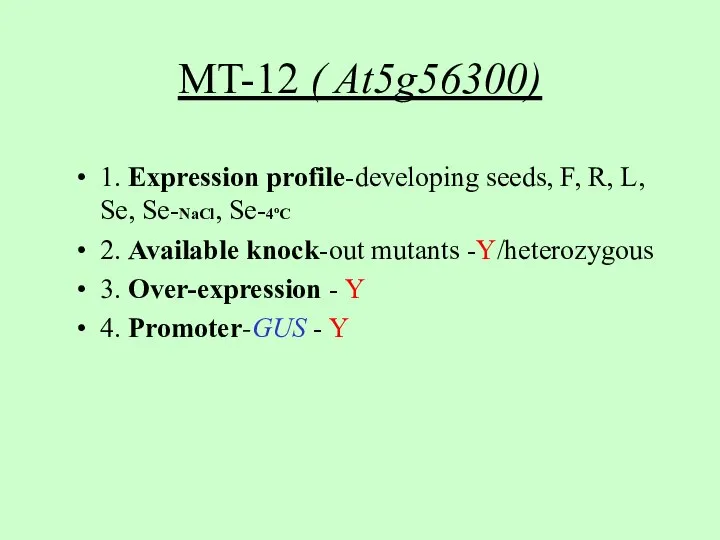
- 8. MT-12 ( At5g56300) 1. Expression profile-developing seeds, F, R, L, Se, Se-NaCl, Se-4oC 2. Available knock-out
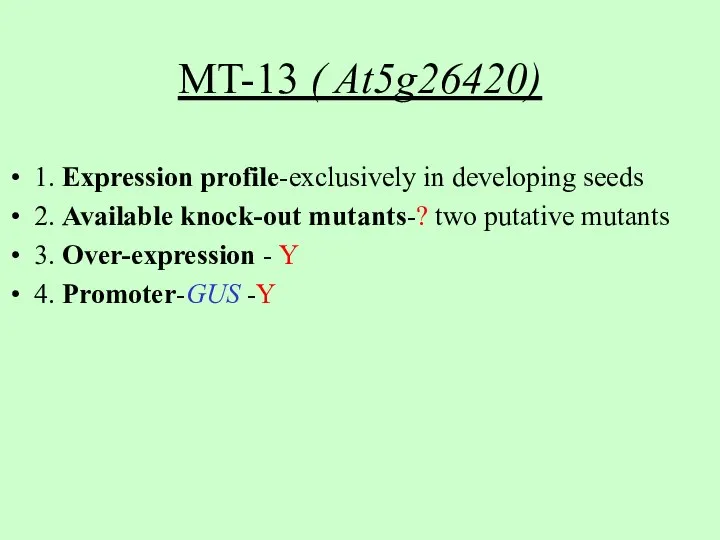
- 9. MT-13 ( At5g26420) 1. Expression profile-exclusively in developing seeds 2. Available knock-out mutants-? two putative mutants
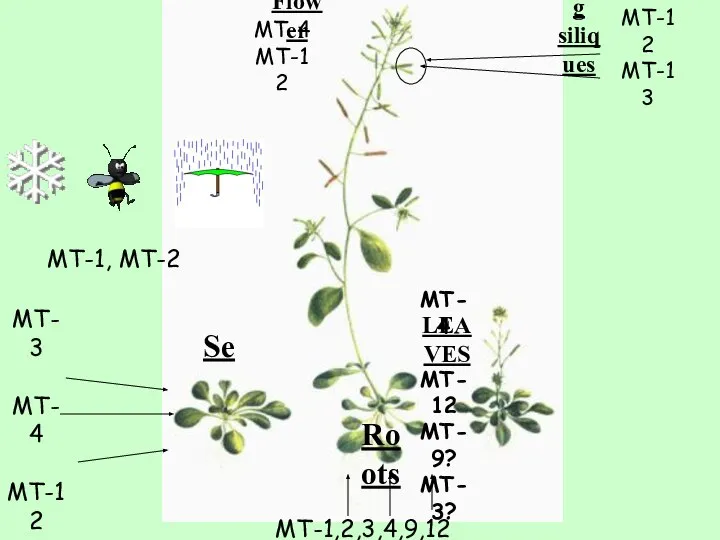
- 10. Back to the first MT-12 MT-13 MT-1,2,3,4,9,12 MT-3 MT-4 MT-12 Se Roots Developing siliques Flower LEAVES
- 12. Скачать презентацию

 Агрессия — инстинктивное поведение животных
Агрессия — инстинктивное поведение животных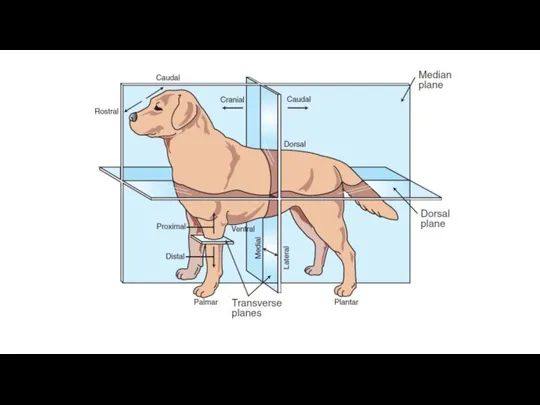 Диапазон движений, возможных в синовиальных суставах
Диапазон движений, возможных в синовиальных суставах Котики – часть нашей жизни
Котики – часть нашей жизни Наследование. Менделевские законы наследования
Наследование. Менделевские законы наследования Биологическая природа человека. Расы человека
Биологическая природа человека. Расы человека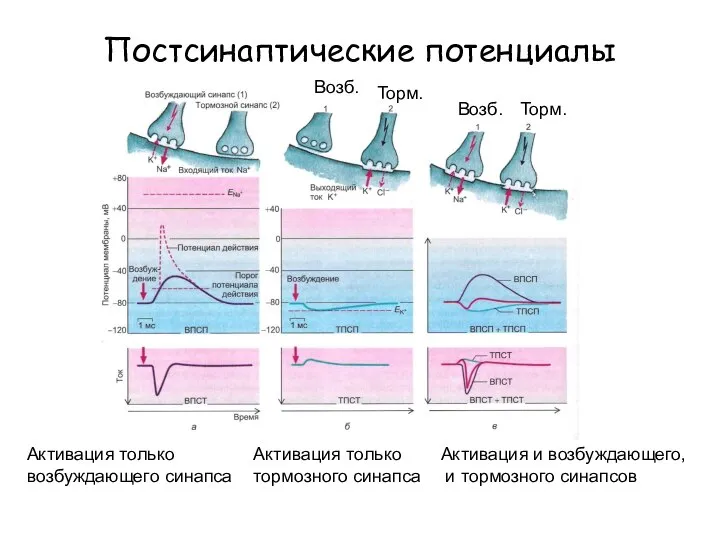 Постсинаптические потенциалы
Постсинаптические потенциалы Презентация на тему ЭКОСИСТЕМА БОЛОТА
Презентация на тему ЭКОСИСТЕМА БОЛОТА  ткани организма человека
ткани организма человека Презентация на тему Трансгенные продукты
Презентация на тему Трансгенные продукты  Игра Угадай-ка
Игра Угадай-ка Теория эволюции по Дарвину
Теория эволюции по Дарвину Приспособленность организмов к среде обитания как результат действия естественного отбора
Приспособленность организмов к среде обитания как результат действия естественного отбора Фармакология процессов обмена веществ и системы крови
Фармакология процессов обмена веществ и системы крови Строение скелета. Кости
Строение скелета. Кости Проект на тему ''Бактерии''
Проект на тему ''Бактерии'' Презентация на тему СЕКРЕТЫ ДОЛГОЛЕТИЯ
Презентация на тему СЕКРЕТЫ ДОЛГОЛЕТИЯ  Лекция 11. Пыльцевая трубка. Основные механизмы полярного роста
Лекция 11. Пыльцевая трубка. Основные механизмы полярного роста Опыт выращивания гороха в разных условиях
Опыт выращивания гороха в разных условиях Ядро клетки
Ядро клетки Бионаноцеллюлоза. Уникальній продукт в практике мировой косметологии
Бионаноцеллюлоза. Уникальній продукт в практике мировой косметологии Тип Плоские черви
Тип Плоские черви Paramyxoviridae
Paramyxoviridae Физиология клетки
Физиология клетки Животные Севера
Животные Севера Анатомия органа зрения
Анатомия органа зрения Турне в огород. Викторина о культурных растениях
Турне в огород. Викторина о культурных растениях Презентация на тему О брюхоногих моллюсках
Презентация на тему О брюхоногих моллюсках  Жизнь птиц в наше время
Жизнь птиц в наше время