Содержание
- 2. Introduction
- 3. Aim and objectives The aim of the work is to reconstruct the phylogenetics, geographic structure and
- 4. The dataset Central Research Institute of Epidemiology kindly provided us with the unique collection of HAV
- 5. Methods Software: MEGA – alignment, tree reconstruction IQTree – model selection BEAST – phylogeography, model selection
- 6. BEAST and VEME2018 The 23rd International Workshop on Virus Evolution and Molecular Epidemiology (VEME2018) was hosted
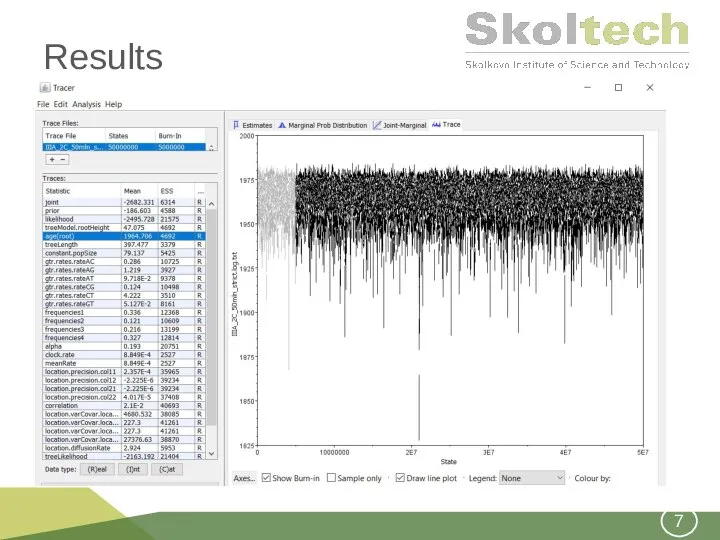
- 7. Results
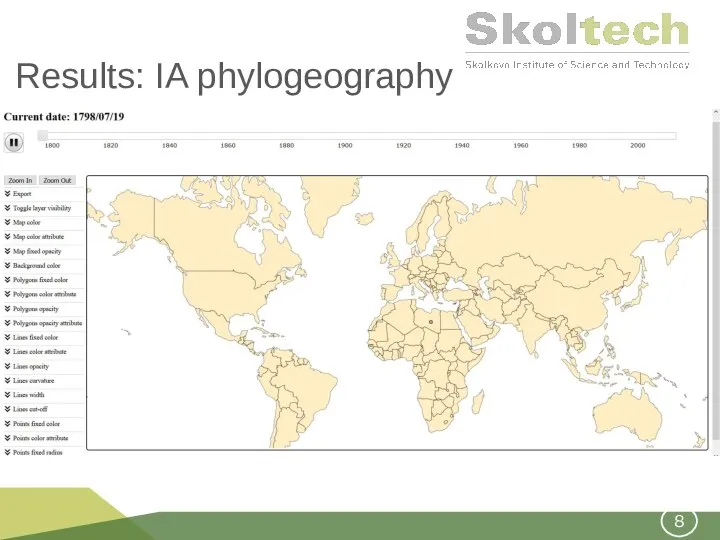
- 8. Results: IA phylogeography
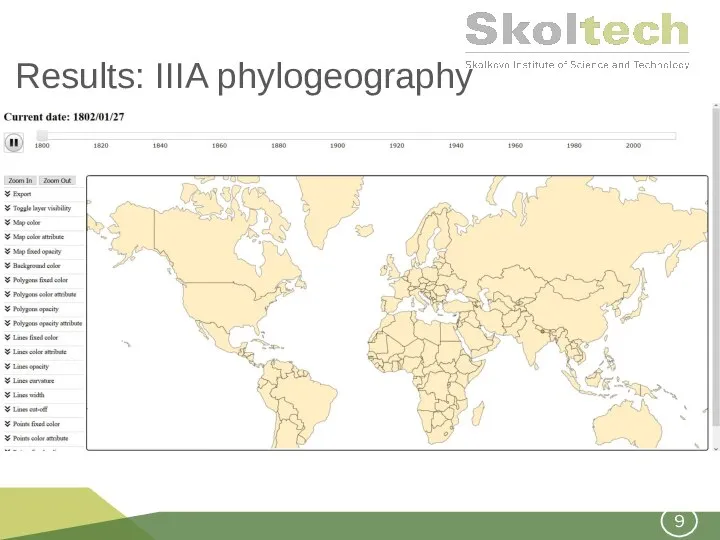
- 9. Results: IIIA phylogeography
- 10. Discussion and problems Sampling issues Uncertainty in time and place of sample collection (current solution –
- 11. Plans Verify BEAST results with other approaches (augur, GenGis) Obtain whole genomes of HAV Analyze the
- 12. Conclusions (obtained, desired) So far, the results of BEAST estimation of phylogeography agree with literature data
- 14. Скачать презентацию



 Школа здоровья для беременных
Школа здоровья для беременных Оказание первой медицинской помощи при получении электротравмы
Оказание первой медицинской помощи при получении электротравмы Климактерический синдром
Климактерический синдром Классификация ХОБЛ. Диспансеризация
Классификация ХОБЛ. Диспансеризация Сердце. Топография сердца. Занятие №1
Сердце. Топография сердца. Занятие №1 БЖА лимфангиомасы және гемангиомасын емдеудің заманауи әдістері
БЖА лимфангиомасы және гемангиомасын емдеудің заманауи әдістері СДР I типа, РДС
СДР I типа, РДС Фитотерапия при заболеваниях ЖКТ
Фитотерапия при заболеваниях ЖКТ №4 Проблемы смерти и умирания в биоэтике. Эвтаназия. Паллиативная медицина
№4 Проблемы смерти и умирания в биоэтике. Эвтаназия. Паллиативная медицина Современные методы инструментальной диагностики заболеваний сердца в гериатрической практике
Современные методы инструментальной диагностики заболеваний сердца в гериатрической практике Дифференциальная диагностика при болях в животе на амбулаторном этапе
Дифференциальная диагностика при болях в животе на амбулаторном этапе Кровотечения из варикозных вен пищевода и желудка
Кровотечения из варикозных вен пищевода и желудка Болезнь туберкулез и ее особенности
Болезнь туберкулез и ее особенности Сопроводительная терапия в онкологии
Сопроводительная терапия в онкологии SPPS терапия
SPPS терапия Люди дождя
Люди дождя Хирургическое лечение тромбофлебита
Хирургическое лечение тромбофлебита Презентация Фитотека
Презентация Фитотека Тест-системы ПЦР в Реальном времени для определения коронавирусной инфекции - компании Гербион ГМБХ
Тест-системы ПЦР в Реальном времени для определения коронавирусной инфекции - компании Гербион ГМБХ Особенности развития костной системы младшего школьника
Особенности развития костной системы младшего школьника Влияние воды на действие лекарственных веществ
Влияние воды на действие лекарственных веществ Экзотические виды массажа
Экзотические виды массажа СПИД- опасное заболевание (8 класс)
СПИД- опасное заболевание (8 класс) Контрастные вещества в рентгенологии
Контрастные вещества в рентгенологии Требования к ситуационной задаче
Требования к ситуационной задаче Качество мяса
Качество мяса Болезнь Лайма. Нейроборрелиоз
Болезнь Лайма. Нейроборрелиоз ВИЧ-инфекция и особенности В-клеточного иммунитета
ВИЧ-инфекция и особенности В-клеточного иммунитета