Содержание
- 2. Reading List Microbial genetics. S.R.Maloy, J.E. Cronan & Freifelder Genetics – a molecular approach. T.A. Brown
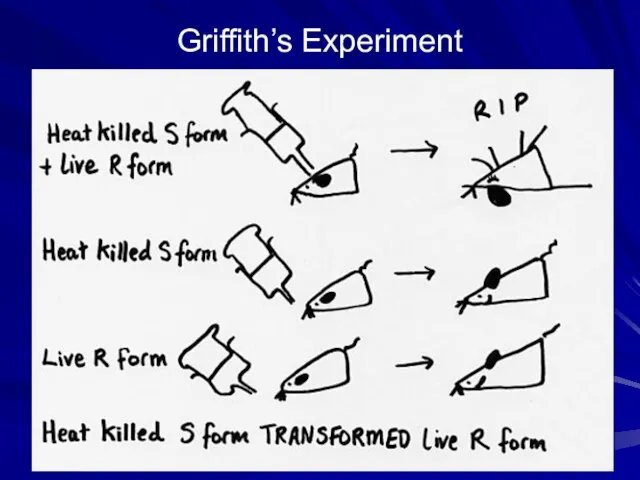
- 3. Bacterial Transformation – uptake of DNA This technique was used to first demonstrate that DNA was
- 4. Griffith’s Experiment
- 5. Transformation Begins with the uptake of DNA chromosomal fragments from the surrounding media into cells competent
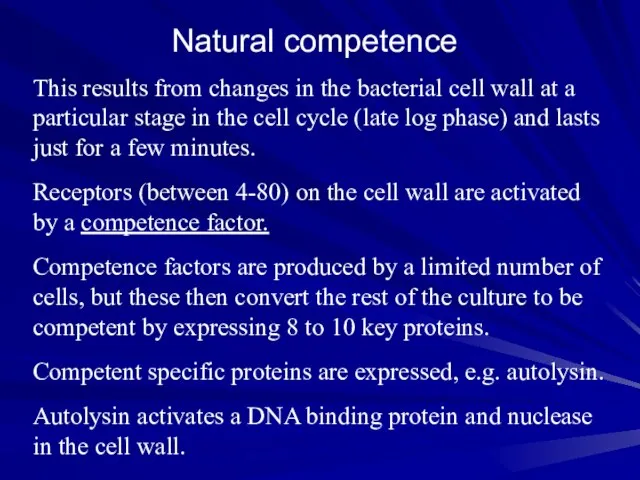
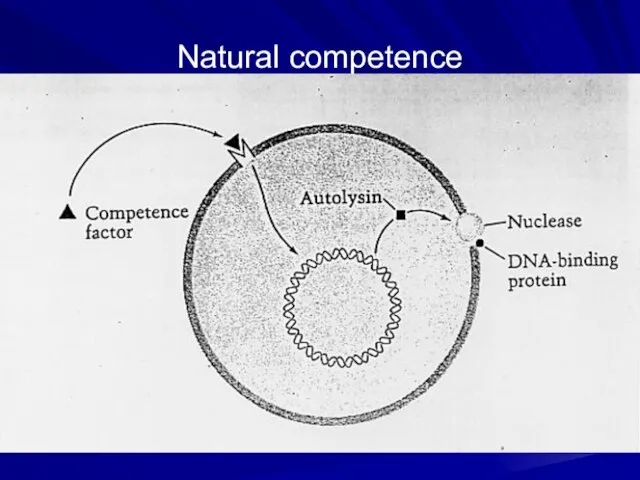
- 6. Natural competence This results from changes in the bacterial cell wall at a particular stage in
- 7. Natural competence
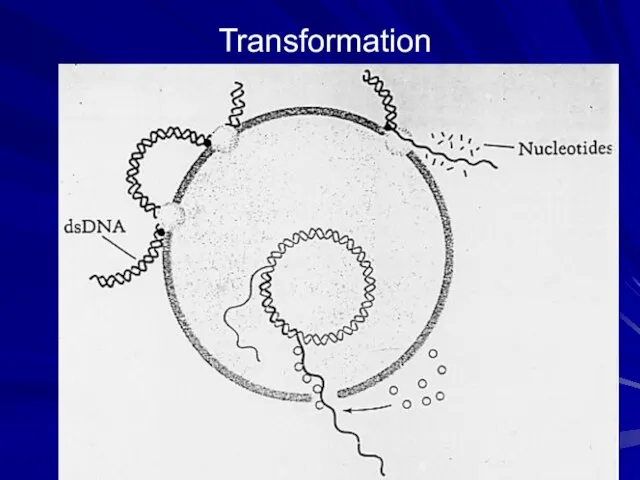
- 8. Transformation cont. Double-stranded DNA (dsDNA) released by bacteria (e.g. during starvation) bind to the ‘competent’ cell
- 9. Transformation
- 10. Competence ‘for uptake of DNA’ Relatively few bacteria are naturally competent, best examples are Bacillus subtilus,
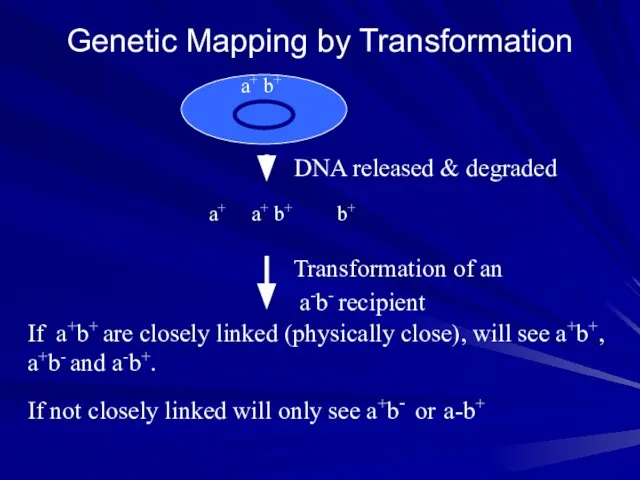
- 11. Genetic Mapping by Transformation a+ b+ b+ a+ Transformation of an a-b- recipient If a+b+ are
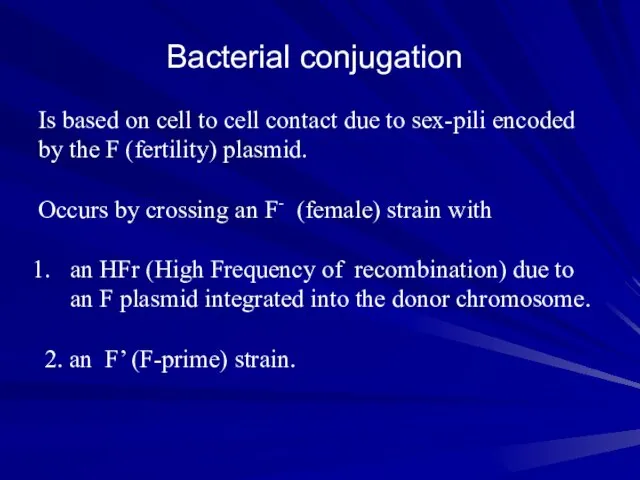
- 12. Bacterial conjugation Is based on cell to cell contact due to sex-pili encoded by the F
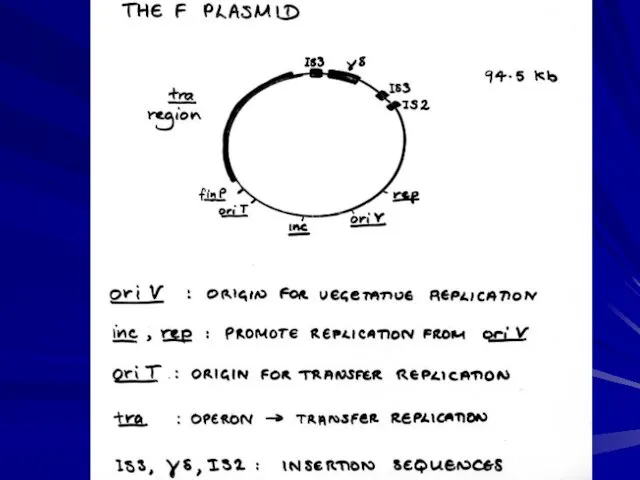
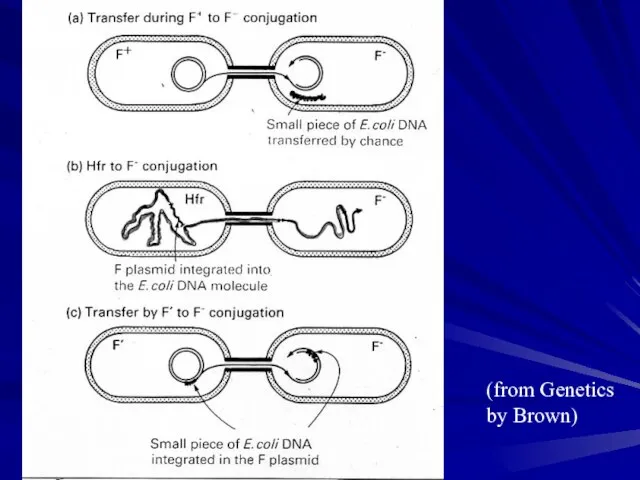
- 14. (from Genetics by Brown)
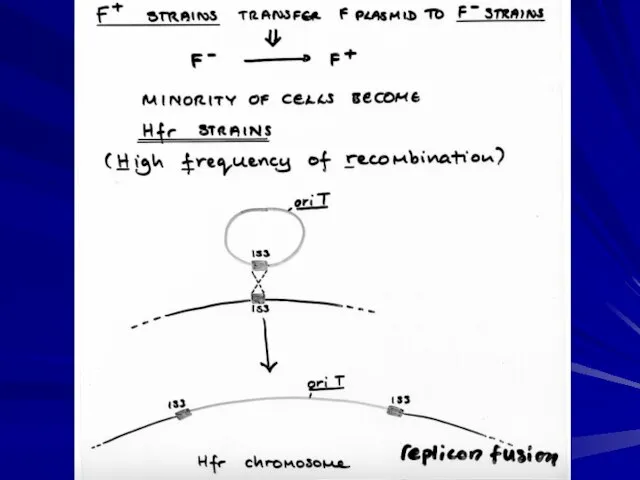
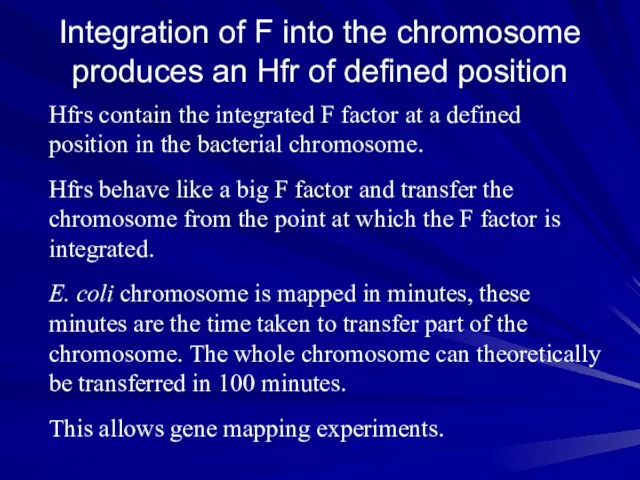
- 16. Integration of F into the chromosome produces an Hfr of defined position Hfrs contain the integrated
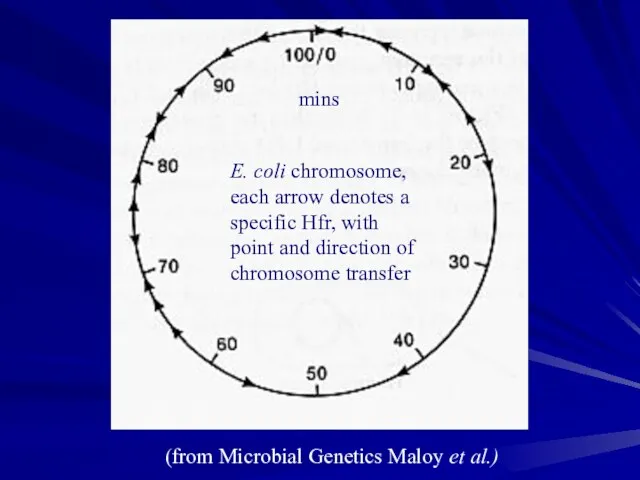
- 17. E. coli chromosome, each arrow denotes a specific Hfr, with point and direction of chromosome transfer
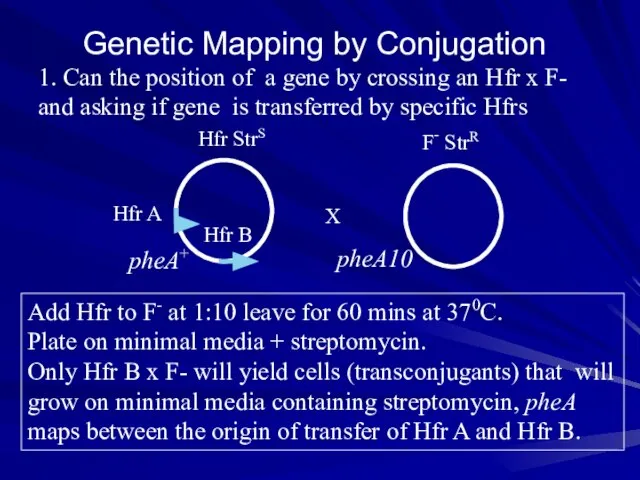
- 18. Genetic Mapping by Conjugation 1. Can the position of a gene by crossing an Hfr x
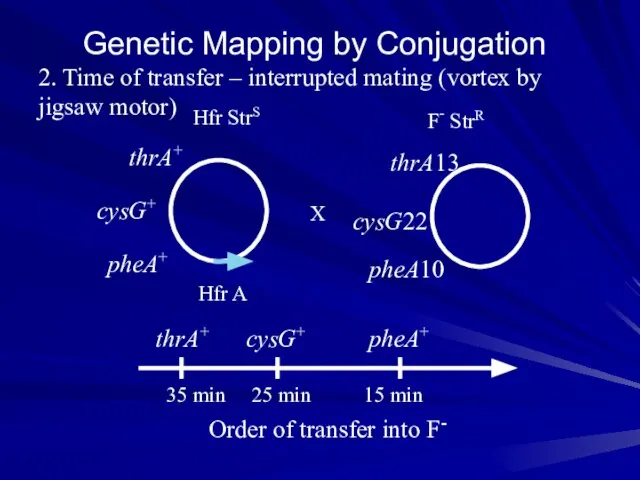
- 19. Genetic Mapping by Conjugation 2. Time of transfer – interrupted mating (vortex by jigsaw motor) Hfr
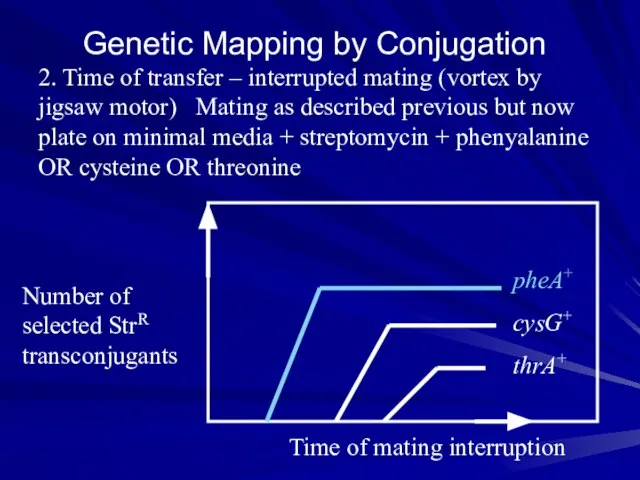
- 20. Genetic Mapping by Conjugation 2. Time of transfer – interrupted mating (vortex by jigsaw motor) Mating
- 22. Скачать презентацию





 Вклад М.В.Ломоносова в русскую литературу
Вклад М.В.Ломоносова в русскую литературу Создание дидактических материалов по истории с использованием средств ИКТ к теме «Занимательная история»
Создание дидактических материалов по истории с использованием средств ИКТ к теме «Занимательная история» О проведении работы по медицинскому обеспечению летней оздоровительной кампании 2012 года в Пермском крае
О проведении работы по медицинскому обеспечению летней оздоровительной кампании 2012 года в Пермском крае Higher education traditions in the USA
Higher education traditions in the USA Великий ученый - энциклопедист
Великий ученый - энциклопедист Типы текста
Типы текста Строим пирамиду
Строим пирамиду Основные элементы оборудования и наполнения среды
Основные элементы оборудования и наполнения среды Презентация на тему Понятие, этапы развития и функции денег
Презентация на тему Понятие, этапы развития и функции денег Оптические иллюзии в живописи и графике
Оптические иллюзии в живописи и графике Культура и общество
Культура и общество Д.Поллок
Д.Поллок BLANCOSUBLINEДизайнерское решение для подстольного монтажа – семейство моек BlancoSubline из Silgranit® CLEAN
BLANCOSUBLINEДизайнерское решение для подстольного монтажа – семейство моек BlancoSubline из Silgranit® CLEAN Топливно-энергетический комплекс мира (ТЭК)
Топливно-энергетический комплекс мира (ТЭК)  Экспериментальная установка теплового насоса
Экспериментальная установка теплового насоса 드라마와 함께 하는 한국어 수 업제2 강강
드라마와 함께 하는 한국어 수 업제2 강강 Сравнение умственной работоспособности обучающихся младшей и старшей школы в стрессовой ситуации
Сравнение умственной работоспособности обучающихся младшей и старшей школы в стрессовой ситуации Условия для реализации основной образовательной программы ДО.
Условия для реализации основной образовательной программы ДО. Школьною тропинкой начинается Жизнь, поверь, у каждого из нас. И о чём мечтали, всё сбывается! Переходим мы из класса в класс…
Школьною тропинкой начинается Жизнь, поверь, у каждого из нас. И о чём мечтали, всё сбывается! Переходим мы из класса в класс…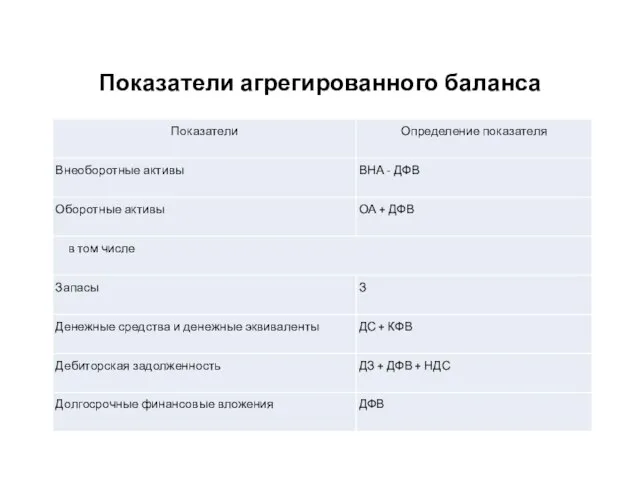 Показатели агрегированного баланса
Показатели агрегированного баланса Сложное предложение (4 класс)
Сложное предложение (4 класс) Презентация Киренский 2022
Презентация Киренский 2022 Самоконтроль в обучении
Самоконтроль в обучении Результаты выполнения Росприроднадзором поручений Минприроды России по подготовке плана по реализации комплекса мер, направленн
Результаты выполнения Росприроднадзором поручений Минприроды России по подготовке плана по реализации комплекса мер, направленн Единственное право и единственная обязанность силы – это защищать слабого Волкодав
Единственное право и единственная обязанность силы – это защищать слабого Волкодав Презентация на тему Древний Вавилон
Презентация на тему Древний Вавилон Школа Панацея 03.09.2020
Школа Панацея 03.09.2020 EUGR NSP CLUB Лекция 3 OMEGA-3 Обзор партнёра компании NSP спортивного врача И.И.Шашкова.
EUGR NSP CLUB Лекция 3 OMEGA-3 Обзор партнёра компании NSP спортивного врача И.И.Шашкова.