Содержание
- 2. Content Intoduction Sructure of SARS-CoV-2 Aim of the study Methods VMD CABS Dock 5. Results
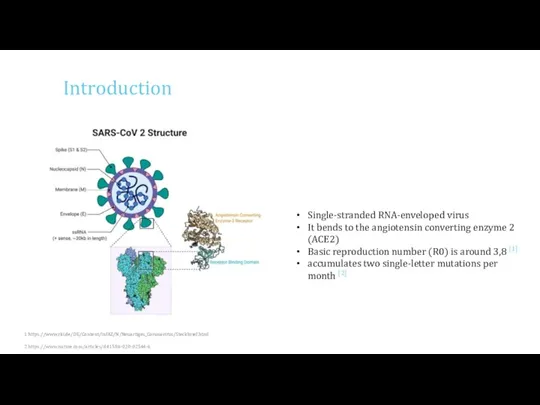
- 3. Introduction 1 https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Steckbrief.html 2 https://www.nature.com/articles/d41586-020-02544-6 Single-stranded RNA-enveloped virus It bends to the angiotensin converting enzyme 2
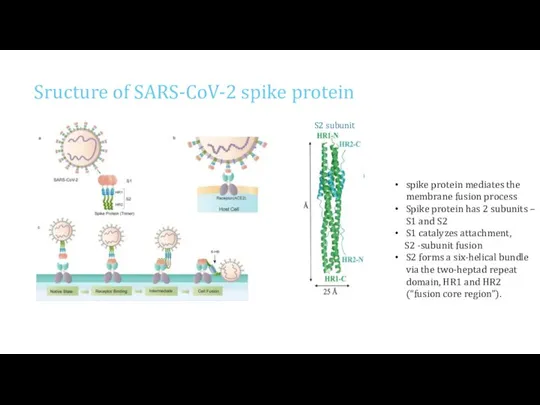
- 4. Sructure of SARS-CoV-2 spike protein spike protein mediates the membrane fusion process Spike protein has 2
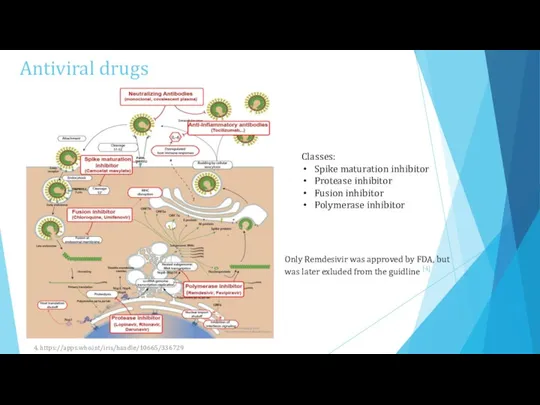
- 5. Antiviral drugs Classes: Spike maturation inhibitor Protease inhibitor Fusion inhibitor Polymerase inhibitor Only Remdesivir was approved
- 7. Aim of the study to screen a list of peptides which were designed to bind the
- 8. Methods
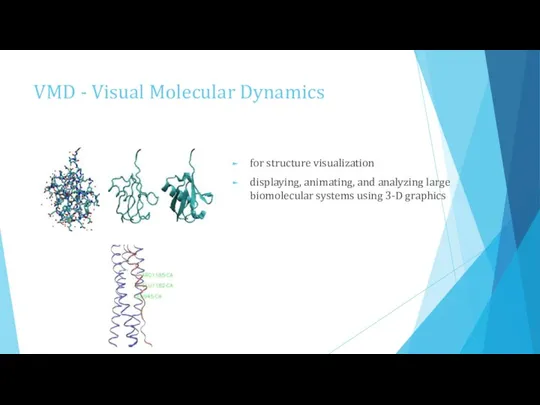
- 9. VMD - Visual Molecular Dynamics for structure visualization displaying, animating, and analyzing large biomolecular systems using
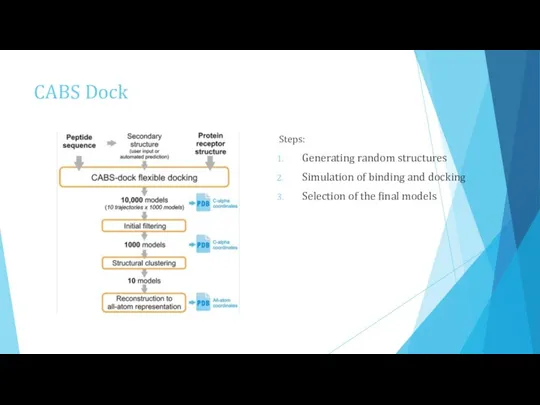
- 10. CABS Dock for protein-peptide docking coarse-grained model (it decreases a time of long simulations) advantages: Can
- 11. CABS Dock Steps: Generating random structures Simulation of binding and docking Selection of the final models
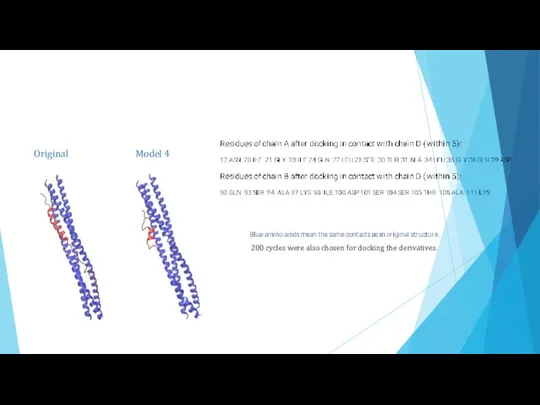
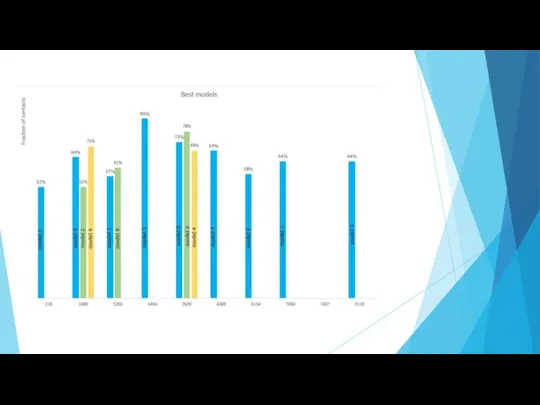
- 12. Results Original structure: Fraction of contacts – 51 % (cutoff – 5Å) number of residues in
- 13. Original Model 4 200 cycles were also chosen for docking the derivatives.
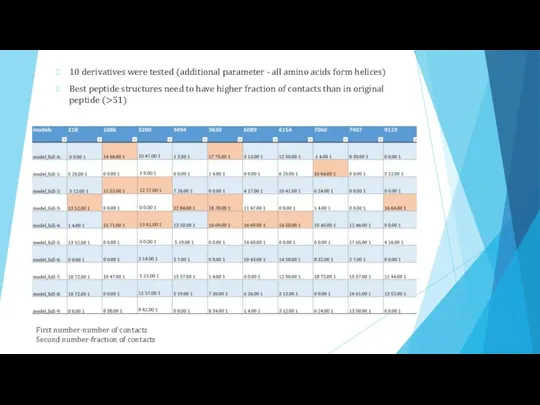
- 14. 10 derivatives were tested (additional parameter - all amino acids form helices) Best peptide structures need
- 16. Ranking according to the predicted number of models, fractions and position of a modal in a
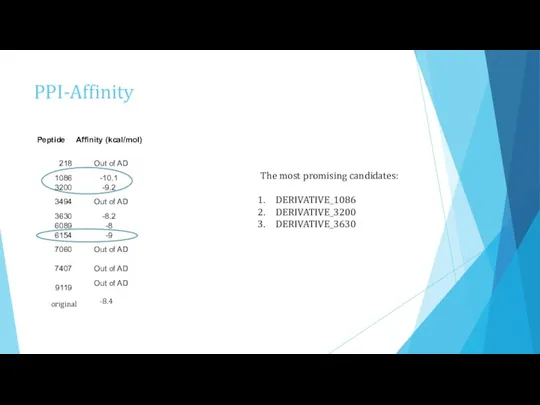
- 17. PPI-Affinity The most promising candidates: DERIVATIVE_1086 DERIVATIVE_3200 DERIVATIVE_3630 original -8.4
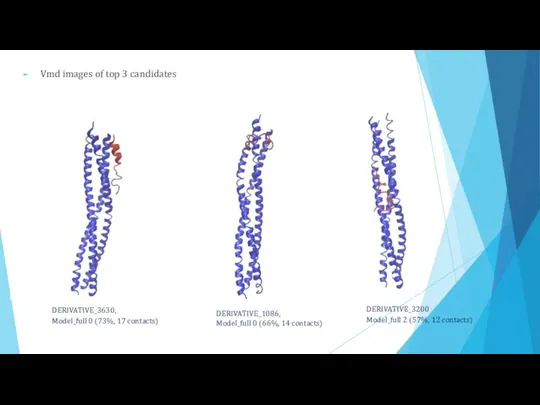
- 18. Vmd images of top 3 candidates DERIVATIVE_3630, Model_full 0 (73%, 17 contacts) DERIVATIVE_1086, Model_full 0 (66%,
- 20. Скачать презентацию






 Специфика послеоперационной работы
Специфика послеоперационной работы Система организации педиатрической помощи
Система организации педиатрической помощи Исследование реактивности коры надпочечников к действию природной минеральной воды курорта Белокуриха
Исследование реактивности коры надпочечников к действию природной минеральной воды курорта Белокуриха Методы обеспечения проходимости дыхательных путей
Методы обеспечения проходимости дыхательных путей Основы синдесмологии
Основы синдесмологии Эмбриогенез нервной системы. Основные этапы
Эмбриогенез нервной системы. Основные этапы Микроэлементы
Микроэлементы Методы обследования в травматологии и ортопедии
Методы обследования в травматологии и ортопедии Применение психотропных препаратов в стоматологии
Применение психотропных препаратов в стоматологии Курение во время беременности и его последствия
Курение во время беременности и его последствия Насыбай
Насыбай Цефалгии: обзор ICHD - 3, мигрень, головная боль напряжения, пучковая головная боль
Цефалгии: обзор ICHD - 3, мигрень, головная боль напряжения, пучковая головная боль Грипп seu Influenza
Грипп seu Influenza Попперс. Как использут попперс
Попперс. Как использут попперс Стандартизация стоматологической продукции
Стандартизация стоматологической продукции Дыхатетельная гимнастика для детей
Дыхатетельная гимнастика для детей Lektsia_5__Khron_alkogolizm
Lektsia_5__Khron_alkogolizm Голова. Отделы головы
Голова. Отделы головы Сот медицинасы және гистология
Сот медицинасы және гистология Черника (лат. Vaccínium myrtíllus)
Черника (лат. Vaccínium myrtíllus) Анатомия и физиология как медицинские науки. Плоскости и оси тела человека
Анатомия и физиология как медицинские науки. Плоскости и оси тела человека Центр разработки нейро-протезов
Центр разработки нейро-протезов Консервативные и оперативные методы лечения гинекологических больных
Консервативные и оперативные методы лечения гинекологических больных Техника торакоскопической резекции пищевода
Техника торакоскопической резекции пищевода Искусственное очищение крови и других жидкостей человеческого тела. Принцип действия диализа. Методы диализа
Искусственное очищение крови и других жидкостей человеческого тела. Принцип действия диализа. Методы диализа Балалардағы асқазанның сәулелік диагностикасы
Балалардағы асқазанның сәулелік диагностикасы Обеспечение соблюдения требований по обращению с медикаментами,
Обеспечение соблюдения требований по обращению с медикаментами, Шизофрения и Психозы шизофренического спектра
Шизофрения и Психозы шизофренического спектра